Utilize R and GBIF to automatically download data and run optimized MaxEnt models for multiple species.
For organizational purposes, I stored the at times lengthy custom functions for this analysis in it's own file.
source("path/to/GBIF_SDM_Functions.R")) You can use the bioclim_download() custom function, or load in your own data layers of interest!
bioclim_download(resolution = 10) # Adjust resolution as neededbioclim_dir = here::here("data", "wc2.1_10m") # Adjust version & resolution path accordingly
bioclim_files = list.files(path = bioclim_dir
, pattern = "\\.tif$", full.names = TRUE)
# Convert to RasterStack for dismo::maxent()
env_rs = raster::stack(bioclim_files)You can create a dataframe or load in your own which contains the scientific name of each species of interest, as well as the country you would like to use for your species' SDM.
# Example Dataframe - Replace W/ Your Own
spp = data.frame(
Scientific.Name = "Adelges tsugae",
Source.Location = "Japan"
)Native ranges for species are typically vague or generalized to an entire region, so we must convert these regions to a list of countries. I created a repository of global regions and their associated countries in order to bridge this gap. Utilize the region_to_country() custom function & region_mapping repository to convert generalized regions to a list of countries. For example, the region of North America would be converted to a vector containing the United States, Canada, and Mexico! This is then converted to country codes to more reliably query GBIF in step 5.
# Convert Country --> Country Code
spp = spp %>%
filter(Source.Location != "")
spp$countryCode = sapply(spp$Source.Location, flexible_country_code)
spp = subset(spp, !is.na(countryCode) & countryCode != "")
# Apply the function to the Source.Location column
spp$Source.Location = sapply(spp$Source.Location, region_to_country, mapping = region_mapping)GBIF is a well respected global effort to document species occurences, and has great integration in R. By utilizing our species names and countries of interest, we can automate the download of point data from the GBIF database! Apply the gbif_occ_data() custom function to all species by their specified locations of interest.
# Apply the function to each scientific name in the dataframe
occ_list = setNames(mapply(gbif_occ_data,
spp$Scientific.Name, spp$countryCode,
SIMPLIFY = FALSE), spp$Scientific.Name)
# Convert the list to a data frame, and also create a new column to hold the Scientific.Name
occ_df = bind_rows(occ_list, .id = "Scientific.Name")
# Remove NA values
occ_df = occ_df[complete.cases(occ_df), ]
# Check how many occurrences subset for each spp.
table(occ_df$Scientific.Name)Maximum Entropy Models (MaxEnt) are a great method to model and predict species distributions based on point data. I have streamlined this process, and incorperated methods to mathematically determine the best setting for each species using the ENMeval package. This package determines the best arguments for your species' dataset such as the beta multiplier and the feature combination. MaxEnt models can be specified by different combinations of five feature combinations:
- L = linear
- Q = quadratic
- H = hinge
- P = product
- T = threshold
ENMeval determines which feature combination is best suited and is setup as inputs for our MaxEnt model. For example, LHT can be a statistically better model choice than just a pure L feature model!
maxent_results = run_maxent(occ_df, env_rs)
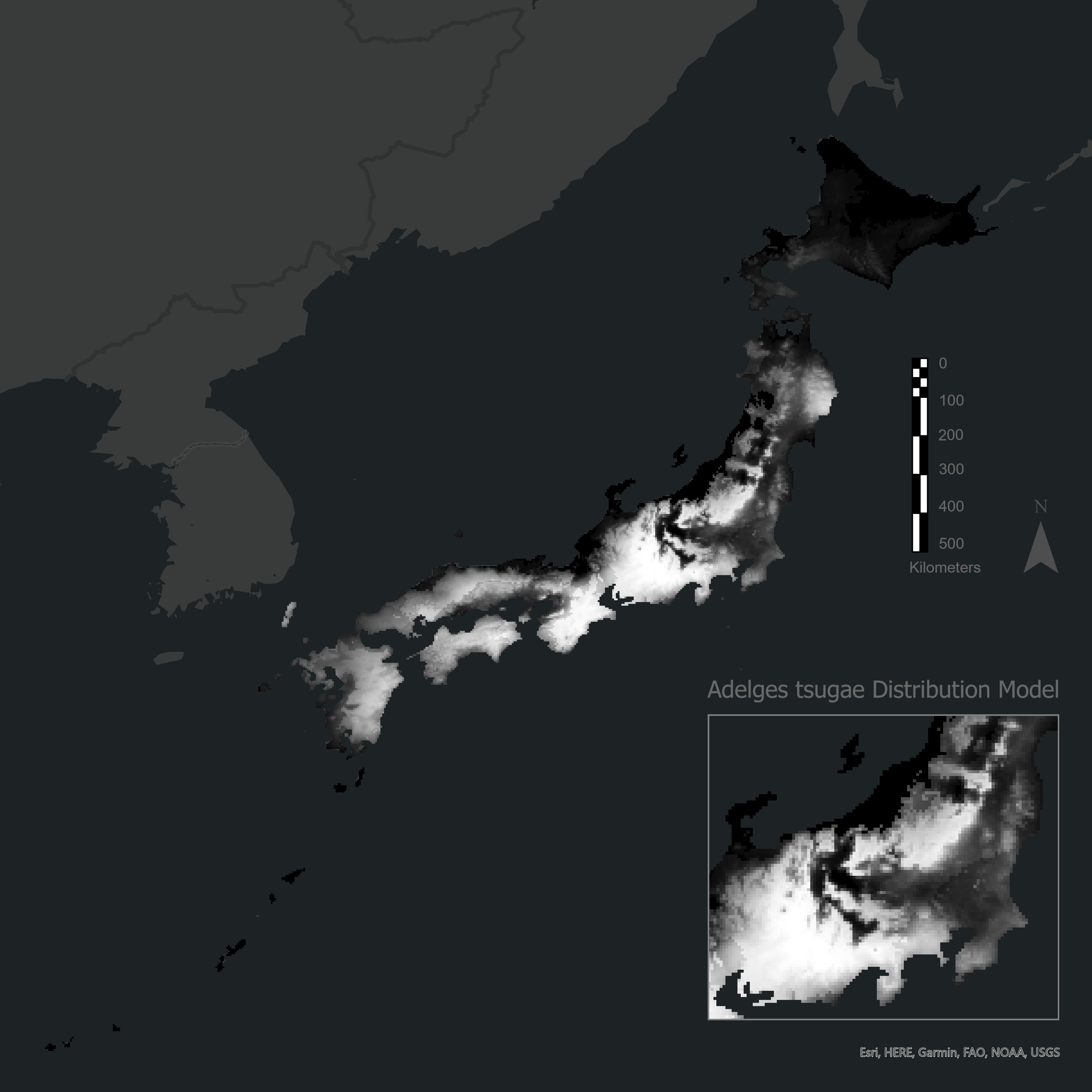
save(maxent_results, file = "maxent_results.rda")Predict a species distribution model for each species in your dataframe by using the complete optimized MaxEnt models!
# Use world country boundaries to clip prediction extent for each species
worldbound = st_read(here::here('data', 'world-administrative-boundaries', 'world-administrative-boundaries.shp'))
# Batch clip prediction extents
clipped_rasters_list <- spp_clip_raster(spp, worldbound, env_rs)
# Create the "MaxEnt_Predictions" directory to store results
dir.create("MaxEnt_Target_Predictions", showWarnings = FALSE)
# Loop through each model and predict
maxent_predict()