By: Erikka Linn, Kalyani Kottilil, Katha Korgaonkar
Intro
Background
What is ATAC-seq?
How Does ATAC-seq Work?
Protocol
Applications
Pros and Cons
Future Developments
References
Current methods such as ChIP-seq, Hi-C and bisulfite sequencing are ways to analyze the effects of epigenetic modifications on the genome. However, these methods require prior knowledge of the mechanisms where one needs to have specific transcription factors (and their subsequent antibodies) available. This limits what potential mutations/changes can be fully investigated. One method that overcomes this limitation is ATAC-seq which can analyze chromatin accessibility no matter the mechanism.
The ATAC-seq protocol only requires around 50,000 cells and two steps: cell preparation and transposition. Open chromatin is tagged with Tn5 transposase and these fragments are then sequenced and analyzed. Applications include nucleosome mapping, transcription factor occupancy analysis, identifying new enhancers, investigating pathological conditions, and single cell analysis to better understand differences between or within cell populations. The speed and efficacy of this procedure makes ATAC-seq optimal to identify open regions of the genome, and future developments are focusing on making the method more high-throughput using microfluidic (and related) techniques.
Chromosomes are condensed DNA where strands are tightly wound and unwound to make the sequences available for regulatory processes. This process is detailed in the figure below:
Transcription factors are proteins that modify DNA and regulate the transcription of genes. This modification requires that the transcription factors are able to physically access and bind to the specific/important areas on the sequence. As such, it is important to be able to locate and find all possible areas that are accessible to regulatory elements since these are likely to be important to biological function. One method that attempts this and was discussed in class is ChIP-seq. However, it is limited to finding and mapping reads that a specific/known transcription factor binds to rather than all possible locations.
Genetic regulation can be carried out through a variety of interactions such as when upstream activation sequences recruit proteins or proximal promoter elements modify strands in addition to the interaction between chromatin and transcription factors.
So, how can the other regulatory regions not modified by transcription factors be found?
This is where ATAC-seq is useful as it can help provide a ‘bigger picture’ where the reads show accessible regions DNA sequences in that relate to specific locations in the nucleosome and binding sites.
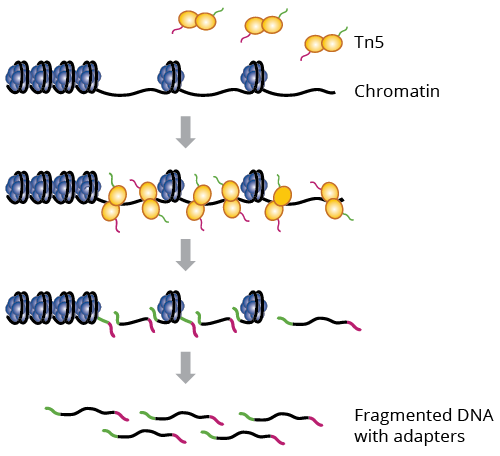
ATAC-seq is an Assay for Transposase-Accessible Chromatin with high throughput sequencing. ATACseq was created at Stanford University in Howard Chang’s and William Greenleaf’s labs. Their goal was to develop a type of chromatin sequencing that would not need as much time and resources as other chromatin sequencing methods. They used the Tn5 transposase because it was common practice and very successful at fragmenting and adding adapters at the same time. They thought that if they used this method on open chromatin, they would be able to discover the chromatin accessibility.
DNA transposons are specific repeating DNA sequences that are “cut & pasted” throughout the genome into open/accessible regions of DNA with the help of a DNA transposase. Tn5 is one such transposase and is commonly used in many experiments. ATAC-seq uses next generation sequencing to explore chromatin accessibility throughout the genome by using a probing hyperactive Tn5 transposase.
The added benefit of this type of transposase is that it is able to fragment and tag regions in the DNA at the same time. This contrasts with most typical sequencing methods which fragment DNA and then tag them afterwards. The result after Tn5 is added DNA are regions fragmented at accessible regions within the genome. From here, the typical sequencing pipeline is carried out where a library is generated, quality control checks are done, and mapping or other further downstream analysis can bring interesting insights.
This protocol only needs around 50,000 cells and a couple hours to produce results. There are two main steps of the protocol: Cell Preparation and Transposition Reaction. In Cell Preparation, cells are collected and counted. Counting the cells is an extremely important step as the count can affect transposition reaction and size distribution of the DNA fragments. The cells are kept in a homogenous solution and are treated with a nonionic detergent to extract the nuclei. In Transposition Reaction, the open chromatin found in the nuclei are tagged with sequencing adapters using Tn5 transposase. After this, the DNA fragments are amplified using PCR and then sequenced using next generations sequencing. Some things to keep in mind are to keep the cell counts from 25,000-75,000. Anything outside these bounds could lower the quality of the data. Furthermore, cutting the chromatin usually results in a lesser quality of the data.
A simple protocol can be completed as follows:
- Harvest Cells
- Lyse cells
- Introduce mutant Tn5 transposases, preloaded with DNA adapters for tagmentation and fragmentation of DNA
- Purify fragmented and tagged DNA
- PCR amplify and purify this amplified DNA
- Sequence and map reads to heterochromatin and euchromatin
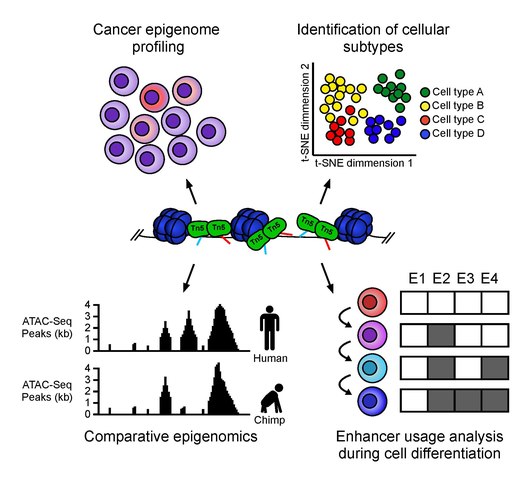
ATAC-seq has a multitude of uses. It can be implemented for things such as nucleosome mapping, transcription factor occupancy analysis, identifying new enhancers, and investigating pathological conditions. It is often used as a way to screen for experiments where researchers can focus follow-up experiments based on changes in chromatin accessibility noticed between samples. For example, ATAC-seq was used to investigate how Polycomb repressive complex 1 (PRC1) affects breast cancer. Furthermore, ATAC-seq has also been used to detect age related diseases such as Age-related macular degeneration (AMD). This procedure uncovered that chromatin accessibility in the retinal regions was extremely decreased in patients with AMD. ATAC-seq can also be used in immunology. Researchers have used it to investigate genes’ role in B-cell development. Finally, ATAC-seq has also been used to study cell differentiation by looking at chromatin accessibility throughout differentiation stages.
Video Explaining how Single Cell ATAC-seq works:
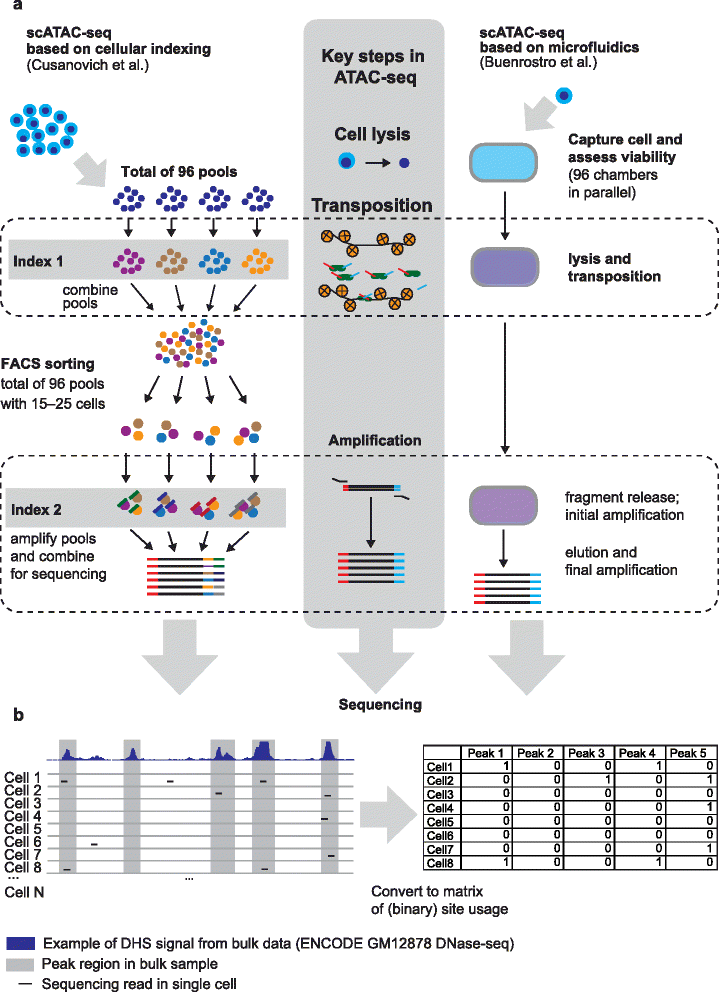
Methods like these tend to average the results from all cell types and therefore single-cell methods are sometimes preferred for certain experiments. ATAC-seq can be modified to include sequencing for single cells. When the chromatin is sheared and tagged with Tn5, each fragment has a unique barcode which can be later on used to identify cell type. snATAC enables researchers to compare chromatin accessibility in different cell types. Analyzing these differences could help in identifying diseases by comparing accessabilities between same cell types in infected vs. non-infected individuals.
| Advantages | Disadvantages |
|---|---|
| 1. ATAC-seq is fast | 1. Not perfect |
| 2. Requires fewer steps | 2. Fragments may be incompatible with PCR |
| 3. High signal-to-noise ratio | 3. Must be optimized from the beginning |
- ATAC-seq is an extremely fast protocol with prep time taking around three hours; it also does not require many cells to run. The simple nature of the library preparation procedure (Tn5 insertion and two rounds of PCR) is what mainly contributes to the speed of the protocol.
- Additional steps such as sonication (used in FAIRE-seq), adding antibodies (done in ChIP-seq), and sensitive enzymatic digestion (used in MNase-seq or DNase-seq) are not required for ATAC-seq, which also contributes to the speed of the protocol. These listed procedures can take up to four days because of the additional steps they require.
- It also has a high signal-to-noise ratio compared to other sequencing methods that measure chromatin accessibility.
- Sometimes chromatin that is bound may sometimes be tagged and sequenced. On average only 50% of the fragments are tagged properly for the PCR amplification step.
- The fragments that do get tagged may not have enough space between the adapters for PCR amplification step.
- The number of cells that the procedure requires must be optimized from the beginning of the protocol. Too few cells could lead to under-transposition, whereas too many cells could lead to over-transposition; samples with too few cells may not yield optimal results.
MNase-seq, DNase-seq, FAIRE-seq, and ATAC-seq are the most commonly used methods to assess primary-order chromatin state.
MNase-seq uses MNase, an endo-exonuclease, to shear linker DNA between nucleosomes, leaving the DNA bound to nucleosomes available to be sequenced. Reads are typically from euchromatic regions, not heterochromatic regions, due to heterochromatin giving rise to longer fragments that can be excluded through size selection before analysis. This method requires 106-107 cells, and at least two biological replicates are needed to make sure the experiment is reproducible. MNase-seq can lend to better understanding of nucleosome positions throughout the genome
DNase-seq uses DNase I exonuclease to sensitively target, digest and subsequently identify open chromatin regions. DNase-seq, like MNase-seq, also requires 106-107 cells and two replicates. The method can be used to also identify binding sites for transcription factors, in addition to finding nucleosome positions.
FAIRE-seq (formaldehyde-assisted isolation of regulatory elements) first crosslinks DNA to nucleosomes using formaldehyde. It then removes the crosslinks and the nucleosome-free DNA is sequenced to ultimately identify open regions in the genome. This method typically does not have high resolution, and the noise-to-signal ratio tends to be high.
It can be observed that ATAC-seq, due to its high resolution, minimal cell requirement, and simplicity, is an optimal technique for analyzing chromatin accessibility.
Future applications or optimizations of this protocol look to apply ATAC-seq in a single cell analysis through microfluidics or combinatorial cellular indexing. With microfluidics, individual nuclei can be isolated and tagmented. Combinatorial cellular indexing circumvents the need to isolate individual cells; it rather uses DNA barcoding to identify open regions of the genome and generate epigenetic profiles. This method does require special technology outside of the scope of what is discussed in this chapter.
Also, due to its ease of use, ATAC-seq is likely to become commonly used in many epigenomics workflows to analyze chromosome accessibility.
- https://www.abcam.com/epigenetics/epigenetics-application-spotlight-atac-seq
- https://www.activemotif.com/blog-atac-seq
- https://www.illumina.com/science/sequencing-method-explorer/kits-and-arrays/atac-seq.html
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. (July 2015). "Single-cell chromatin accessibility reveals principles of regulatory variation". Nature. 523 (7561): 486–90. Bibcode:2015Natur.523..486B. doi:10.1038/nature14590. PMC 4685948. PMID 26083756.
- Buenrostro, Jason D et al. “ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide.” Current protocols in molecular biology vol. 109 21.29.1-21.29.9. 5 Jan. 2015, doi:10.1002/0471142727.mb2129s109
- Chan, Ho Lam, et al. “Polycomb Complexes Associate with Enhancers and Promote Oncogenic Transcriptional Programs in Cancer through Multiple Mechanisms.” Nature Communications, vol. 9, no. 1, 2018, doi:10.1038/s41467-018-05728-x.
- Chang, Pearl et al. “Computational Methods for Assessing Chromatin Hierarchy.” Computational and structural biotechnology journal vol. 16 43-53. 15 Feb. 2018, doi:10.1016/j.csbj.2018.02.003
- Hatzi, Katerina, et al. “Histone Demethylase LSD1 Is Required for Germinal Center Formation and BCL6-Driven Lymphomagenesis.” Nature Immunology, vol. 20, no. 1, 2018, pp. 86–96., doi:10.1038/s41590-018-0273-1.
- Klemm, S.L., Shipony, Z. & Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20, 207–220 (2019) doi:10.1038/s41576-018-0089-8
- Lara-Astiaso, D., et al. “Chromatin State Dynamics during Blood Formation.” Science, vol. 345, no. 6199, 2014, pp. 943–949., doi:10.1126/science.1256271.
- Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, Pokholok D, Aryee MJ, et al. (2019). "Droplet-based combinatorial indexing for massive scale single-cell epigenomics". bioRxiv. doi:10.1101/612713
- Sun, Yuanyuan, et al. “Detect Accessible Chromatin Using ATAC-Sequencing, from Principle to Applications.” Hereditas, vol. 156, no. 1, 2019, doi:10.1186/s41065-019-0105-9.
- Wang, Jie, et al. “ATAC-Seq Analysis Reveals a Widespread Decrease of Chromatin Accessibility in Age-Related Macular Degeneration.” Nature Communications, vol. 9, no. 1, 2018, doi:10.1038/s41467-018-03856-y.