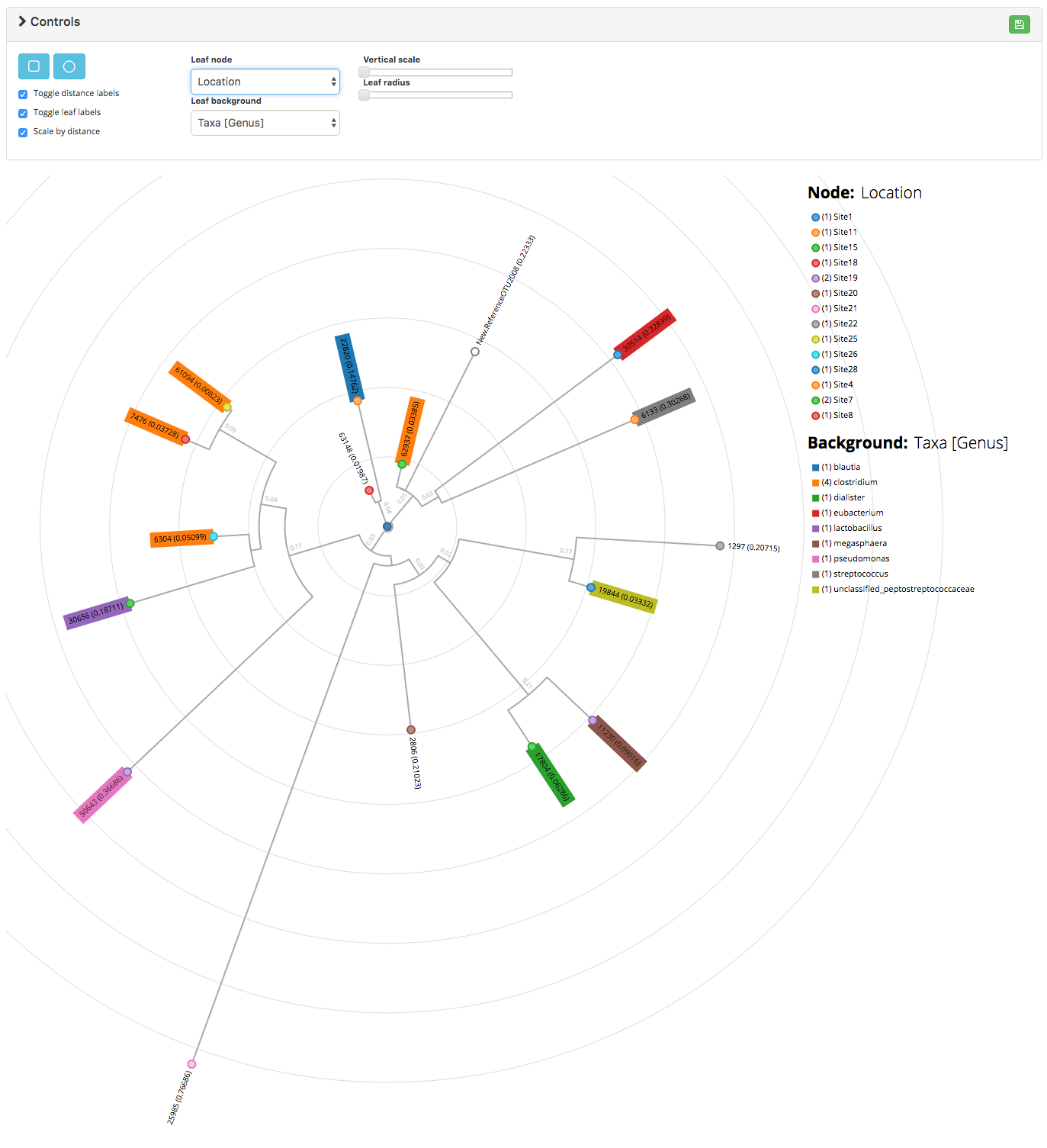
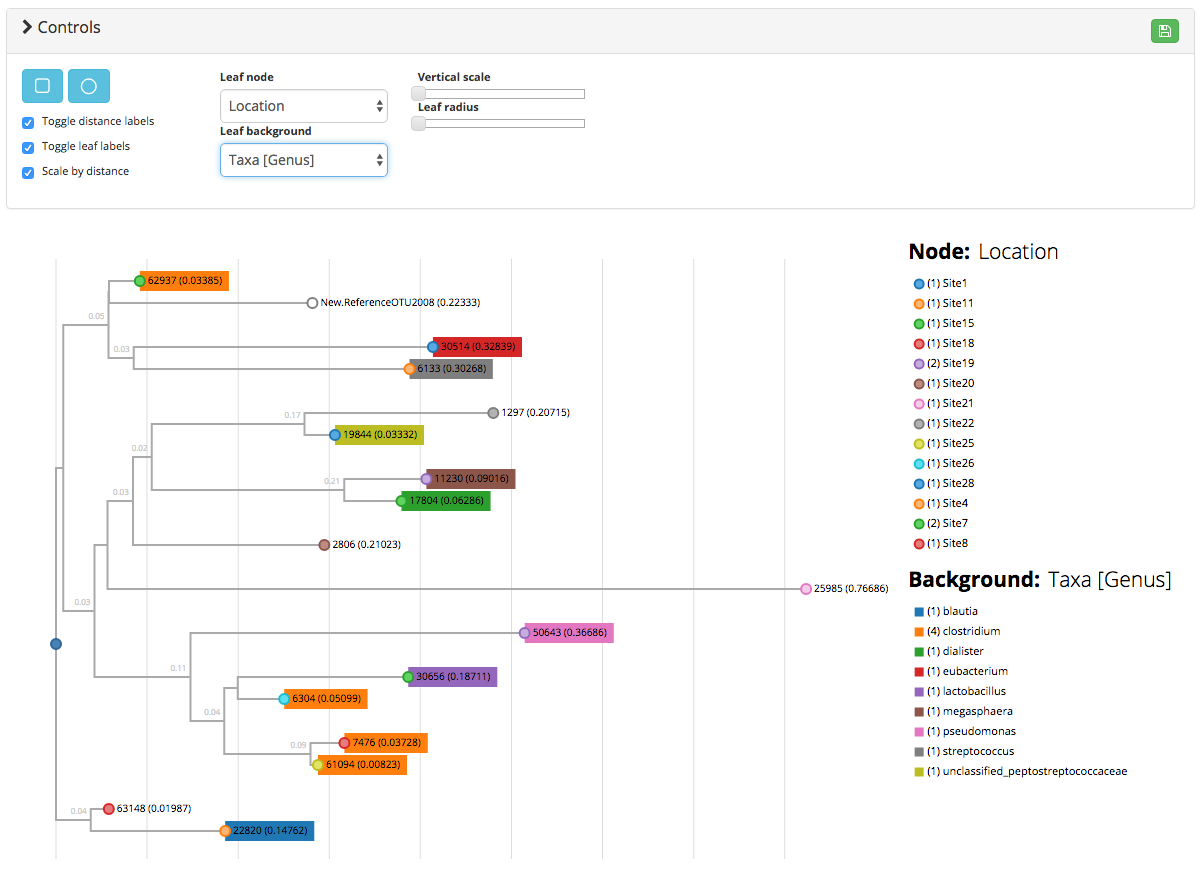
This project is an adaptation of the GitHubGist created by kueda. Given a Newick formatted file as well as an OTU mapping file containing metadata for the leaves in the tree, the script is used to generate a rooted phylogenetic tree:
A phylogenetic tree or evolutionary tree is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities—their phylogeny—based upon similarities and differences in their physical or genetic characteristics. The taxa joined together in the tree are implied to have descended from a common ancestor.
A index.html file has been provided to render the tree as an example.
A working demo can be found here which was used to generate the following:
in development
Rendering the phylogram begins with the function init() which you can simply call in the body tag of your HTML page using the onload event handle: <body onload="init(parameters);">. Currently, the function takes three parameters:
-
String of the Newick tree used to build the phylogram.
-
String identifier for the div id into which to generate the phylogram, note that it must contain the '#' in it. For example, if you want to render your tree in a div with the id 'phylogram' you'd pass '#phylogram' to the function and you'd need the following in your HTML:
<div id='phylogram'></div>. -
an optional object with tree parameters, this should be passed as a javascript object with the following optional keys
- mapping_dat :
(obj)list of objects where the object keys are metadata and the object values are the metadata values. This is used to color the phylogram. - treeType :
(str)either rectangular or radial [default: rectangular] - hideRuler :
(bool)if True, the background ruler will be hidden [default: show ruler] - skipBranchLengthScaling :
(bool)if True, a cladogram will be displayed instead of a phylogram [default: phylogram] - skipLabels :
(bool)if True, all node labels will be hidden [default: show labels]
The mapping_dat is similar to a standard QIIME mapping file in that it associates additional metadata to the objects of interest; however in this case the objects of interest are OTUs instead of samples. This file has the following caveats:
-
each object must have a key of
OTUIDwhich must exist as a leaf in the Newick tree -
if providing taxonomy data, format it in the QIIME default manner (e.g. k__bacteria_2;p__firmicutes;c__negativicutes;o__selenomonadales;f__veillonellaceae;g__megasphaera;s__elsdenii). If done correctly, this format will be parsed and cleaned up automatically
-
if using categorical data (e.g. GroupA, GroupB) the legend will render a row for every category value
-
if passing ordinal data (e.g. 0.4, 1.90) the legend will render a color bar with min/max values. Therefore, if you have categorical data labeled with numbers (e.g. Site 1, 2, 3), ensure that you format the values with alphanumeric characters (e.g. Site1, Site2, Site3)