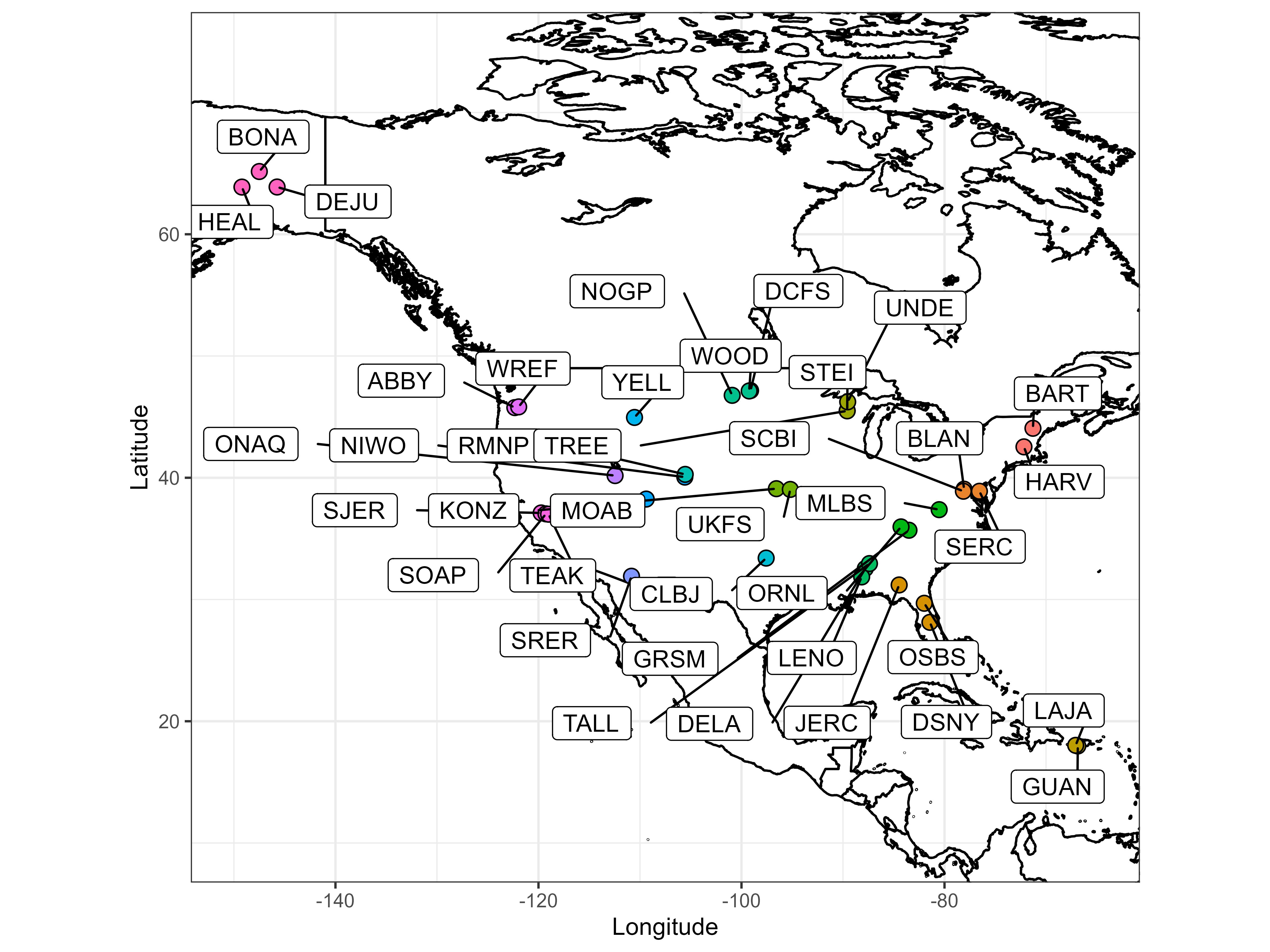
The NEONForestAGB data product includes aboveground biomass (AGB) estimates from individual tree diameters scaled to whole tree biomass using generalized allometric equations for 35 National Ecological Observatory Network (NEON) sites within the United States and Puerto Rico. The data set includes 93,971 unique individuals of 478 different species in 1,216 terrestrial observation plots for 245,245 biomass estimates between the years 2014 to 2023 (current as of July 1, 2024).
To bring the data directly into the R workspace, first download or copy the file ./R/NEONForestAGB.R from this repo--ideally into a project folder and then call that using a relative path †.
Direct URL: https://github.com/atkinsjeff/NEONForestAGB/blob/main/R/NEONForestAGB.R
Then the following code will directly import the NEONForestAGB data into the workspace:
# Change this to wherever you download the file
source("./R/NEONForestAGB.R")
# import the data
df <- getAGB()
Metadata can be viewed and downloaded via getAGBMeta() and the source taxonomy and allometry data can be viewed and downloaded bia getAGBTaxons().
Alternatively, one can copy the function contents of the NEONForestAGB.R file or access the data directly via the figshare repository.
† If you haven't read Jenny Bryan's post on project oriented workflows, please do so! https://www.tidyverse.org/blog/2017/12/workflow-vs-script/
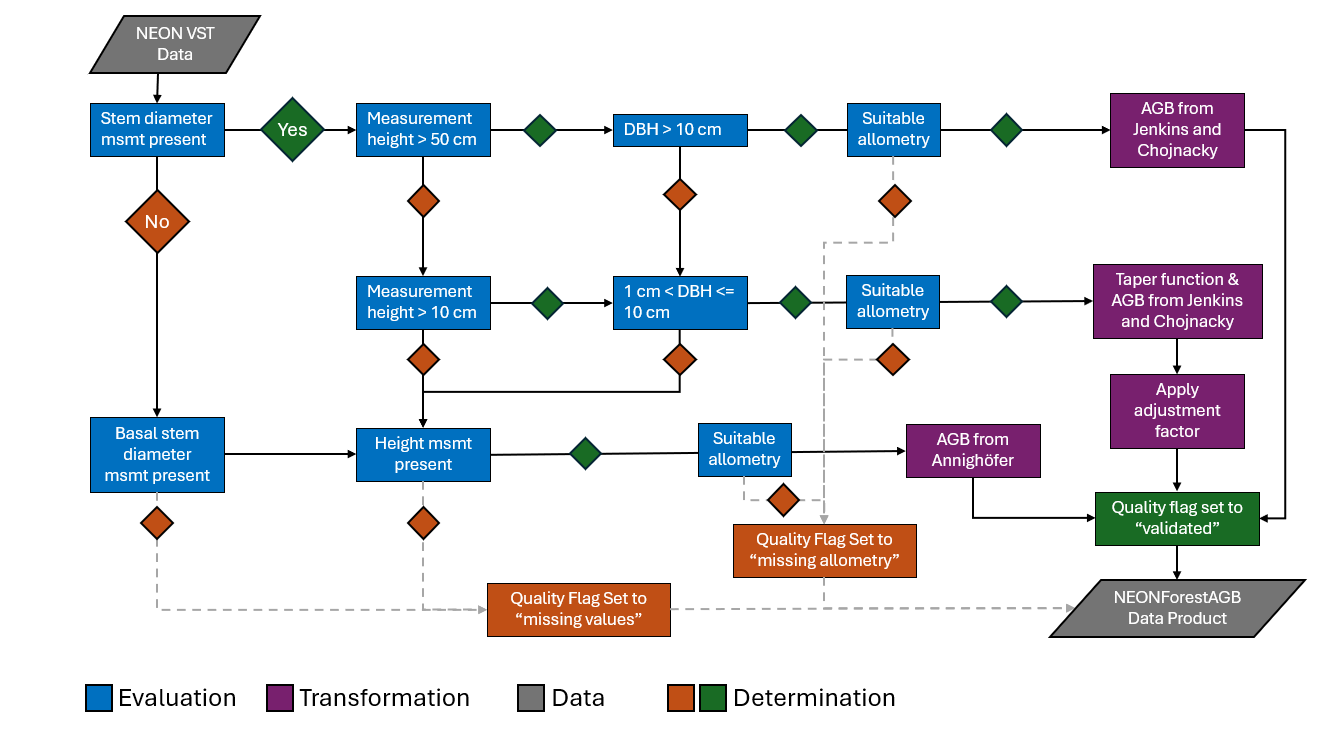
This code uses the Jenkins et al. 2003 and Chojnacky et al. 2014 allometric equations fit to NEON taxon ID information for 40 NEON terrestrial sites to generate estimate of total AGB for individuals.
The Jenkins allometric equations classify trees into several groups (e.g., mixed hardwood, pine, woodland) with a specific equation of the form:
Where
Individual trees in the NEON data set are assigned a taxonID which was used to match with the assigned groups from Jenkins et al. 2003 and Chojnacky et al. (2014).
However, where there was no initially obvious classification group that could be assigned, the USDA PLANTS database and other sources were consulted and a best guess was made. Please feel free to offer suggestions for improvements.
Taxon classification information is found in the ./data/MasterTaxonList.csv file.
Source data files used in the creation of this data product are included in the ./R/ directory. Of particular interest would be the ./R/001_allometry_script_20240320.R file which demonstrates how the data are imported using the neonstore package and then merged with allometric and taxonomy source data we compiled to determine biomass. With proper citation, this file may be adapted and used for your own purposes.
All derived data products, code, and scripts are supported and covered by a CC0 1.0 Universal license. https://creativecommons.org/publicdomain/zero/1.0/legalcode.txt
Jeff W. Atkins (USDA Forest Service, Southern Research Station); Brandon Alveshere (Virginia Commonwealth University); Lu Zhai (Oklahoma State University); Courtney Meier (Battelle-NEON); Michael Langley (USDA Forest Service, Southern Research Station); Sabrie Breland (USDA Forest Service, USFS-Savannah River)
Atkins, Jeff; Meier, Courtney; Alveshere, Brandon; Breland, Sabrie; Langley, Michael (2024). NEONForestAGBv2. figshare. Dataset. https://doi.org/10.6084/m9.figshare.25625352.v1
Chojnacky, D. C., Heath, L. S., & Jenkins, J. C. (2014). Updated generalized biomass equations for North American tree species. Forestry, 87(1), 129-151.
Jenkins, J. C., Chojnacky, D. C., Heath, L. S., & Birdsey, R. A. (2003). National-scale biomass estimators for United States tree species. Forest science, 49(1), 12-35.
National Ecological Observatory Network (NEON). (2021). Woody plant vegetation structure (DP1.10098.001): RELEASE-2021 (RELEASE-2021, p. 624.3 MB) [csv]. National Ecological Observatory Network (NEON). https://doi.org/10.48443/E3QN-XW47
NEON (2023). NEON.DOC.000987-TOS Protocol and Procedure: VST Measurement of Vegetation Structure. https://data.neonscience.org/documents/10179/1883155/NEON.DOC.000987vM/4ff8197 e-3858-6cd8-e3f7-6babf96b2482
Ter-Mikaelian, M. T., and M. D. Korzukhin. 1997. Biomass equations for sixty-five North American tree species. Forest Ecology and Management 97:1--24.
USDA Plants Database. (2024). . https://plants.usda.gov/home.
Woudenberg, S. W., B. L. Conkling, B. M. O'Connell, E. B. LaPoint, J. A. Turner, and K. L. Waddell. 2010. The Forest Inventory and Analysis Database: Database description and users manual version 4.0 for Phase 2. Gen. Tech. Rep. RMRS-GTR-245. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 336 p. 245.