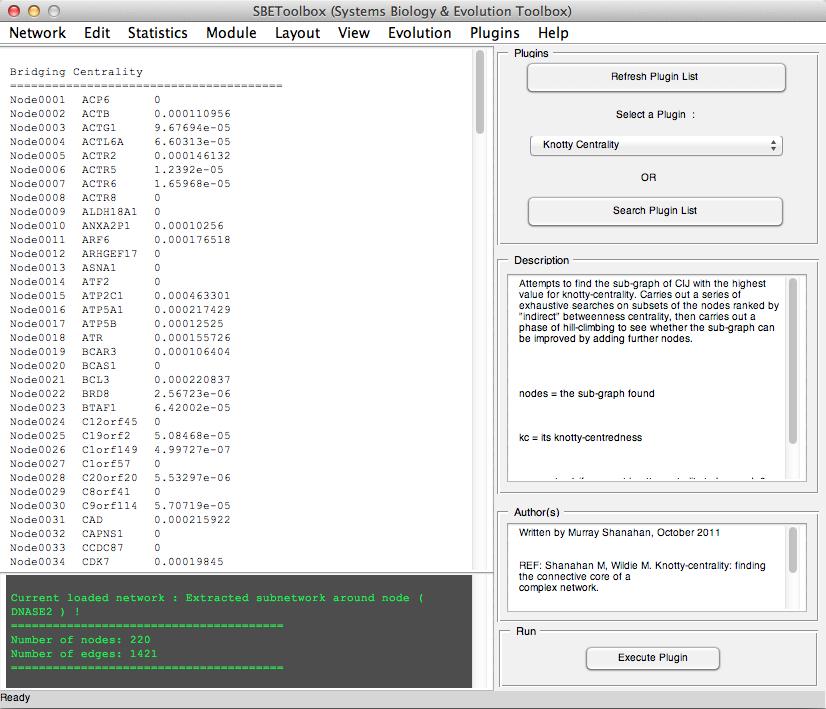
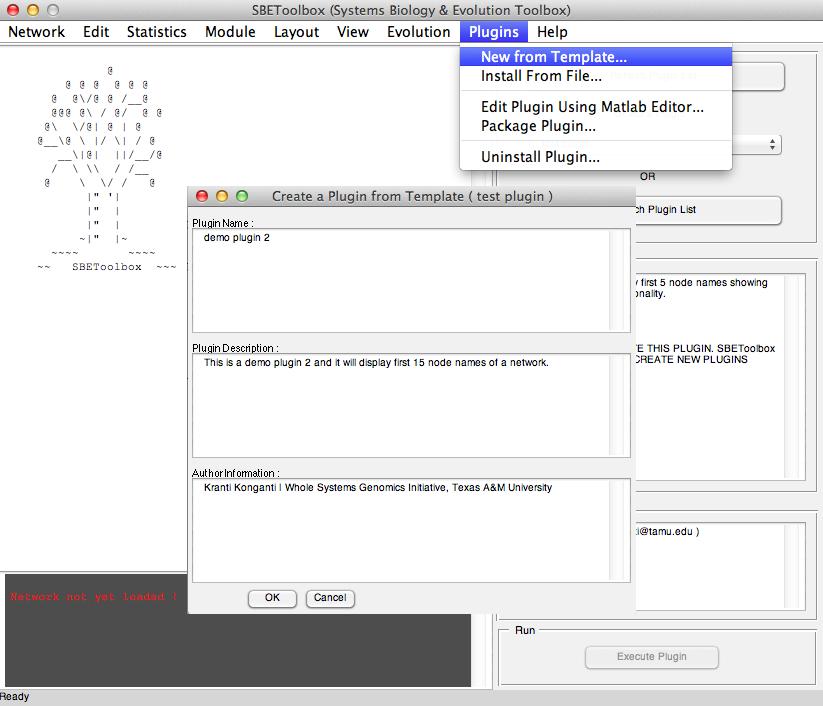
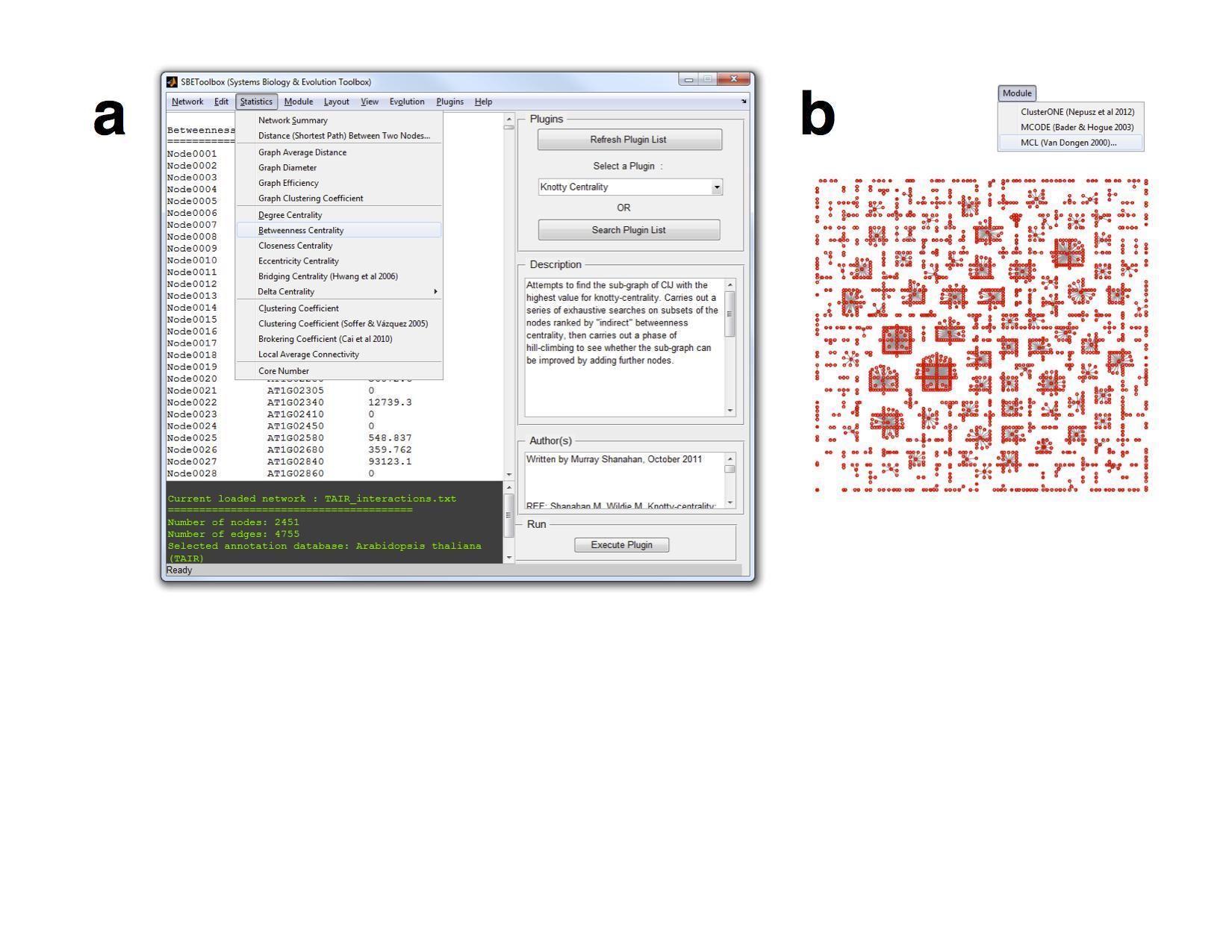
SBEToolbox (Systems Biology and Evolution Toolbox) is developed in MATLAB as a menu-driven UI software to determine various statistics of the biological network.
Some of its features include (but not limited to): algorithms to create random networks (small-world, ring lattice etc..), deduce clusters in the network (MCL, mCode, clusterOne) etc...
Short WIKI: https://promis.tamu.edu/projects/sbetoolbox/wiki/SBEToolbox_User_Manual.
Some Screencasts: http://vimeo.com/64151952 and http://vimeo.com/63437114.
Contact us on Twitter (@SBEToolbox).
Konganti K, Wang G, Yang E, Cai JJ (2013). SBEToolbox: a Matlab Toolbox for Biological Network Analysis.* Evolutionary Bioinformatics 2013:9 355-362