This repository contains code and figures related to the paper On the Spectrum of Finite Rooted Homogeneous Trees joint with my Ph.D. advisor Dan Rockmore. The main theorems in the paper show that for trees with regular branching properties the adjacency spectrum can be realized as the roots of a recursively defined sequence of polynomials and the the associated spectral measures converge to singular distributions.
The current version contains Sage code for three basic tasks: constructing trees, computing their eigenvalues, and displaying the eigenvalue plots. There is also code for generating simplicial versions of these objects and an @interact tool for computing the endpoints of the limiting distribution for regular trees. In order to run the code online you can copy the scripts directly to my Sage cell here: link .
The original versions of these tools were developed for exploratory purposes - more polished versions are in development.
Embedded version: here
This is an interact module for constructing regularly branching trees and computing their eigenvalues. The parameters are the number of branches or nodes adjacent to the root and a list representing the branching pattern. Here is an example of the interface and results for the (\alpha\beta) trees discussed in Theorem 8:
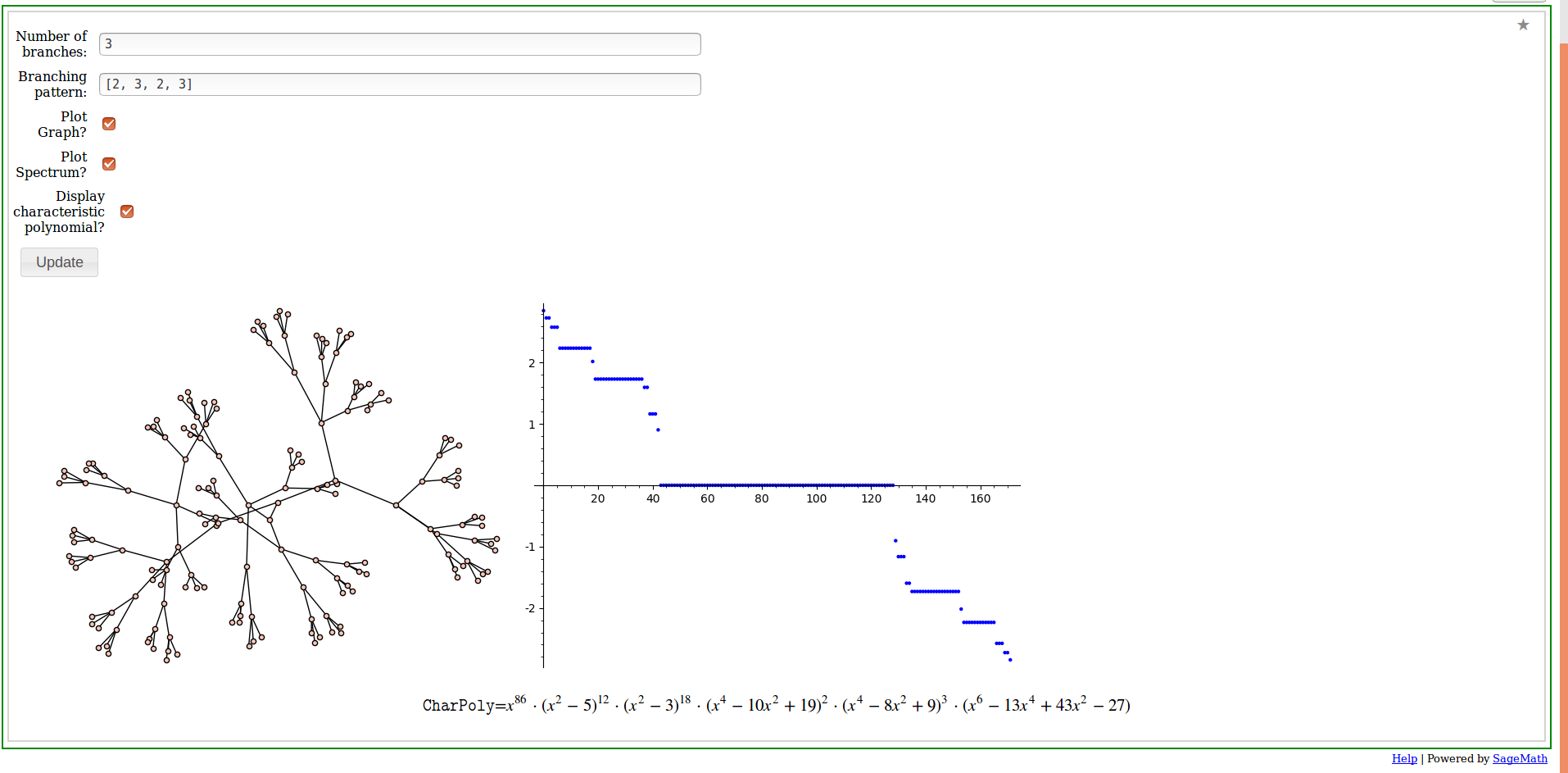
The un-normalized versions of the eigenvalue plots in Figure 2 can be generated by setting the values in the branching pattern list to all be equal.
Embedded version: here
This is an interact module for computing the endpoints of the singular distributions as described in Theorem 6. The two parameters are the branching pattern of the tree (k) and the level of the tree (j). That is, the endpoints correspond to the new eigenvalues that appear in Xjk. The function returns a list of the corresponding left-endpoints and the intervals have width equal to 2-(k+1) by Lemma 6 so the right endpoints can be computed from this information as well.
This contains a collection of individual functions for constructing 3, 4, or 5 regular trees and computing/plotting their full adjacency spectrum.
This contains the constructor for the rooted fans (simplicial complexes) that are discussed in the future work section. The code introduces a recursive labelling scheme to identify the individual 0-cells and their adjacency.
This contains code for computing the upper adjacency graphs for the edges in the rooted fans, as well as the associated spectral information.
This is a catchall file with the experimental code that was used to carry out this research.
Numerical verification tool for Theorem 9. Notice that when a_n = n this is just the statement that \lim_{n\rightarrow\infty} \frac{n!-(n-1)!}{\sum_{k=1}^n k!} = 1