MassVision is a software solution developed in 3D Slicer platform for end-to-end analysis of Mass Spectrometry Imaging (MSI) data, particularly Desorption ElectroSpray Ionization (DESI) modality.
The functionalities include data exploration via various targeted and non-targeted visualization, co-localization to spatial labels (histopathology annotations), dataset generation with spatial- and spectral-guidance, multi-slide data aggregation via feature alignment, denoising via spatial aggregation, machine learning model training, and whole-slide model deployment.
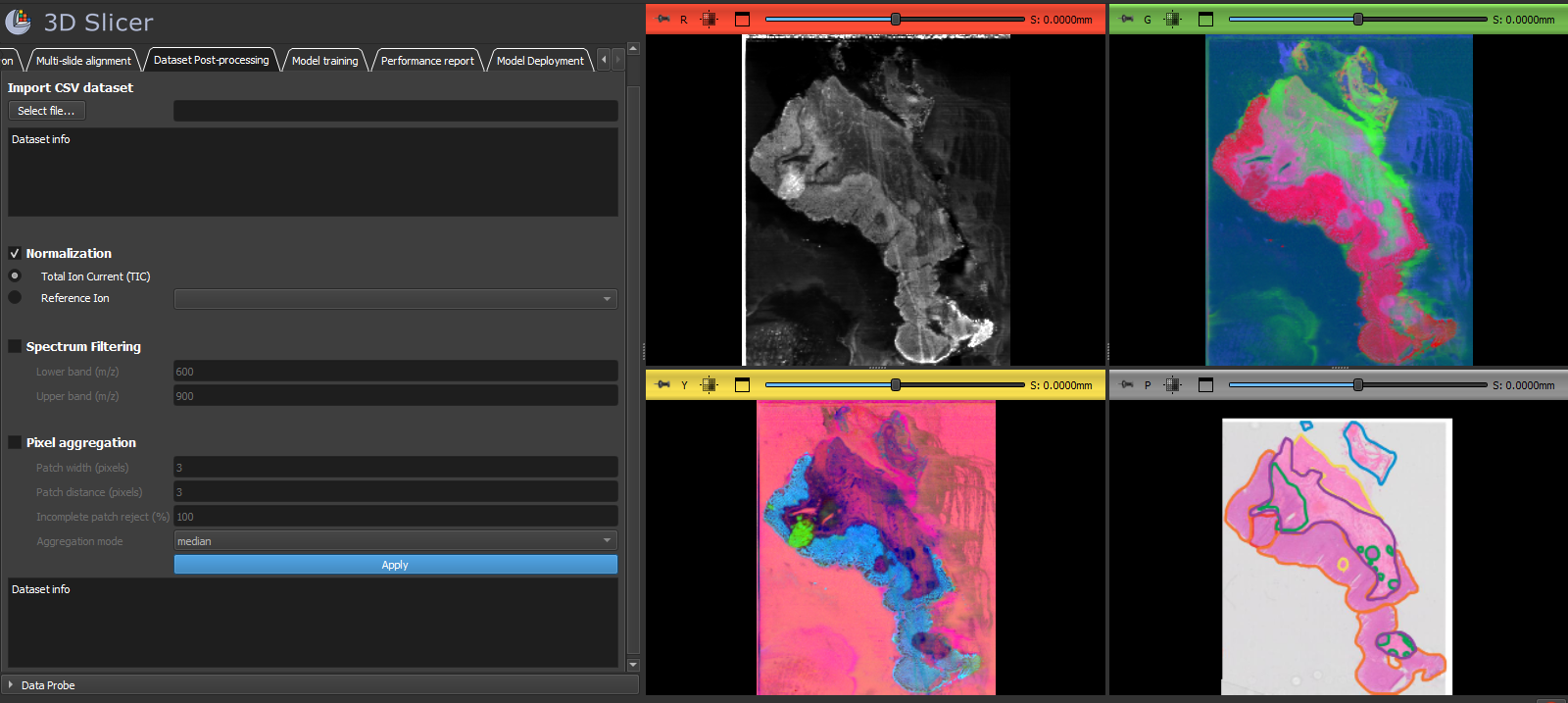
- Data format: DESI TXT images, structured CSV MSI
- Visualization: targeted (single-ion heatmap, multi-ion colormap), untargeted (global PCA, regional PCA, pixel spectrum)
- Dataset generation: spatial colocalization to pathological annotation, labelled ROI extraction
- Multi-slide peak alignment and dataset merge
- Preprocessing: normalization (TIC, single-ion), subband selection, spatial pixel aggregation
- Model training: data stratification, data balancing, model selection
- Whole-slide deployment: global deployment, masked deployment
- Visualization with global vs local contrast demo
- Spatial colocalization of mass spectrometry visualization with histopathology annotations demo
- ROI selection with pathology guide for dataset generation demo
Click here to access user manual, documentations, and tutorials
Please use the following citations if you are using MassVision
- TBA