This repo implements the point process model of neural sequences (PP-Seq) in PyTorch. The original model is described in:
Alex H. Williams ☕, Anthony Degleris 🌄, Yixin Wang, Scott W. Linderman 📢.
Point process models for sequence detection in high-dimensional neural spike trains.
Neural Information Processing Systems 2020, Vancouver, CA.
Here, we implement a simpler inference algorithm that works in discrete time, and a PyTorch implementation takes advantage of parallelization across time bins using a GPU.
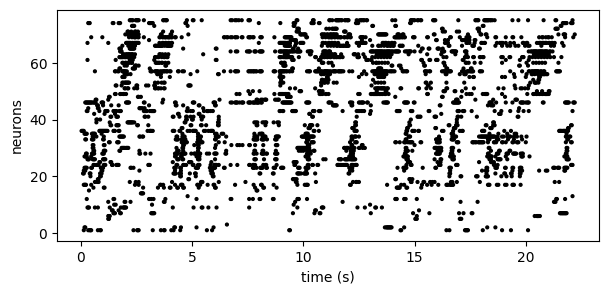
This model aims to identify sequential firing patterns in neural spike trains in an unsupervised manner. For example, consider the spike train below(1):
By eye, we see no obvious structure in these data. However, by re-ordering the neurons according to PP-Seq's inferred sequences, we obtain:
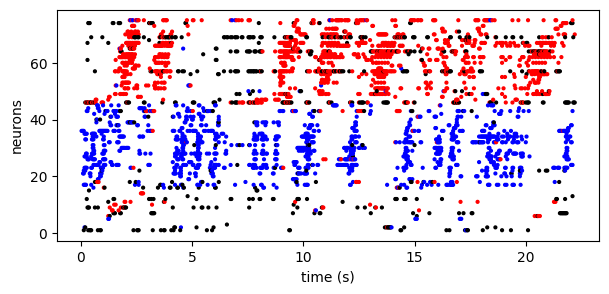
Further, the model provides (probabilistic) assignment labels to each spike. In this case, we fit a model with two types of sequences. Below we use the model to color each spike as red (sequence 1), blue (sequence 2), or black (non-sequence background spike):
Footnote. (1) These data are deconvolved spikes from a calcium imaging recording from zebra finch HVC. These data were published in Mackevicius*, Bahle*, et al. (2019) and are freely available online at https://github.com/FeeLab/seqNMF.