tesliper: Theoretical Spectroscopist's Little Helper is a program for batch processing
of Gaussian output files, focused on calculations of vibrational, electronic, and
scattering spectra from Gaussian-calculated quantum properties of molecule's conformers.
It allows to easily exclude conformers that are not suitable for further analysis:
erroneous, not optimized, of higher energy or lower contribution than a user-given
threshold. It also implements an RMSD sieve, enabling one to filter out similar
structures. It lets you calculate theoretical IR, VCD, UV, ECD, Raman, and ROA spectra
for each conformer or as a population-weighted average and export obtained spectral data
in one of supported file formats: .txt, .csv, or .xlsx. Finally, if allows to
easily setup a next calculations step with batch export to .gjf Gaussian input files.
tesliper is written in Python 3.6 and makes use of some additional third party
packages (see below or requirements.txt). It may be used as a package or as a
stand-alone application with dedicated GUI.
You can use tesliper from python or as standalone application with dedicated graphical
user interface. See below for details.
Python 3.6+
numpy
openpyxl
matplotlib (optional, for GUI)
tesliper uses tkinter to deliver the graphical interface. It is included in
most Python distributions, but please be aware, that some might miss it. You will
need to install it manually in such case.
tesliper is available on PyPI, you can install it to your python distribution by
simply running:
$ python -m pip install tesliper
or
$ python -m pip install tesliper[gui]
if you would like to be able to use a graphical interface.
This option is currently available only for Windows users.
To get your copy of tesliper, simply download a Tesliper.exe file from the
latest relase.
This file is a standalone application, no installation is required.
Full documentation is available online: https://tesliper.readthedocs.io/.
Conventions that are important to note:
tesliperstores multiple data entries of various types for each conformer. To prevent confusion with Python's datatypeand with data itself,tesliperrefers to specific kinds of data as "genres". Genres in code are represented by specific strings, used as identifiers. To learn about data genres known totesliper, see Available data genres.tesliperidentifies conformers using a stem of an extracted file (i.e. its filename without extension). When files with identical names are extracted in course of subsequentTesliper.extractcalls or in recursive extraction usingTesliper.extract(recursive=True), they are treated as data for one conformer. This enables to join data from subsequent calculations steps, e.g. geometry optimization, vibrational spectra simulation, and electronic spectra simulation. Please note that if specific data genre is available from more than one calculation job, only recently extracted values will be stored.tesliperwas designed to deal with multiple conformers of single molecule and may not work properly when used to process data concerning different molecules (i.e. having different number of atoms, different number of degrees of freedom, etc.). If you want to use it for such purpose anyway, you may setTesliper.conformers.allow_data_inconsistencytoTrue.tesliperwill then stop complaining and try to do its best.
Tesliper class is the main access point to tesliper's functionality. It allows you
to extract data from specified files, provides a proxy to the trimming
functionality, gives access to data in form of specialized arrays, enables you
to calculate and average desired spectra, and provides an easy way to export data.
Most basic use might look like this:
from tesliper import Tesliper
tslr = Tesliper()
tslr.extract()
tslr.calculate_spectra()
tslr.average_spectra()
tslr.export_averaged()This extracts data from files in the current working directory, calculates available spectra using standard parameters, averages them using available energy values, and exports to current working directory in .txt format.
You can customize this process by specifying call parameters for used methods
and modifying Tesliper's configuration attributes:
- to change source directory
or location of exported files instantiate
Tesliperobject withinput_dirandoutput_dirparameters specified, respectively. You can also set appropriate attributes on the instance directly; - to extract only selected files in
input_dirusewanted_filesinit parameter. It should be given an iterable of filenames you want to parse. Again, you can also directly set an identically named attribute; - use
Tesliper.conformers.trim...methods to easily filter out conformers you wish to ignore in further analysis; - to change parameters used for calculation of spectra, modify appropriate entries
of
parametersattribute; - use other export methods to export more data and specify
fmtparameter in method's call to export to other file formats.
tslr = Tesliper(input_dir="./myjob/optimization/", output_dir="./myjob/output/")
tslr.wanted_files = ["one", "two", "three"] # only files with these names
tslr.extract() # use tslr.input_dir as source
tslr.extract(path="./myjob/vcd_sim/") # use other input_dir
tslr.conformers.trim_not_optimized() # filtering out unwanted conformers
tslr.parameters["vcd"].update({"start": 500, "stop": 2500, "width": 2})
tslr.calculate_spectra(genres=["vcd"]) # we want only VCD spectrum
tslr.average_spectra()
tslr.export_averaged(mode="w") # overwrite previously exported files
tslr.export_activities(fmt="csv") # save activities for analysis elsewhere
tslr.output_dir = "./myjob/ecd_sim/"
tslr.export_job_file( # prepare files for next step of calculations
route="# td=(singlets,nstates=80) B3LYP/Def2TZVP"
)If you are using tesliper as a standalone application, simply double click on the
Tesliper.exe file to start the application. To invoke it from the command line,
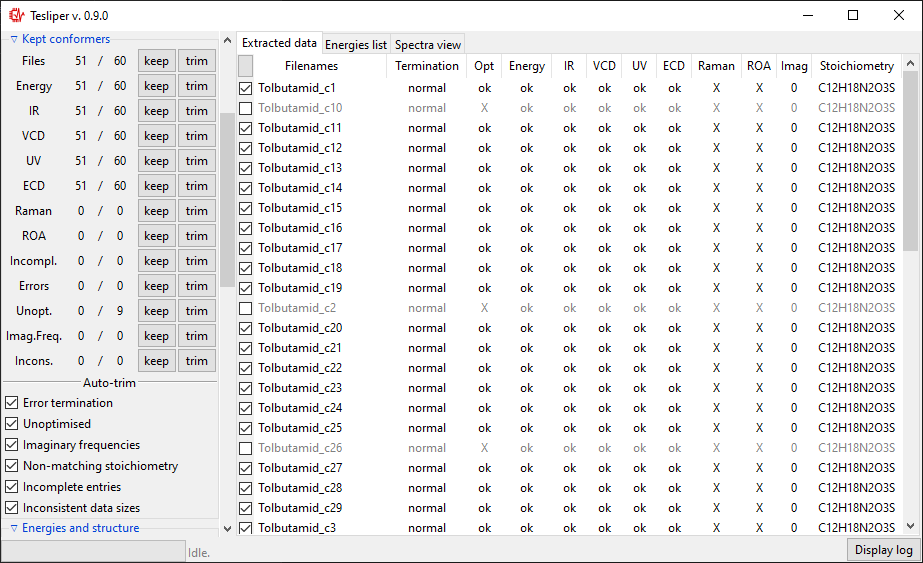
just run tesliper-gui. GUI consists of three panels and a number of controls.
The panels are: "Extracted data", "Energies list", and "Spectra view". First two
offer a list of conformers read so far using "Chose files" and "Chose folder" buttons
on the left. The last enables to preview calculated spectra.
- "Extracted data" panel shows an identifier of each conformer (a file name) and an overview of data extracted. Little checkboxes on the left of each conformer may be clicked to mark this conformer as "kept" or "not kept".
- "Energies list" offers the same set of conformers and checkboxes, but with energies values listed for each conformer. The view may be changed using "Show" dropdown box in "Energies and Structure" section of controls, to present difference in energy between conformers or their percentage contribution in population.
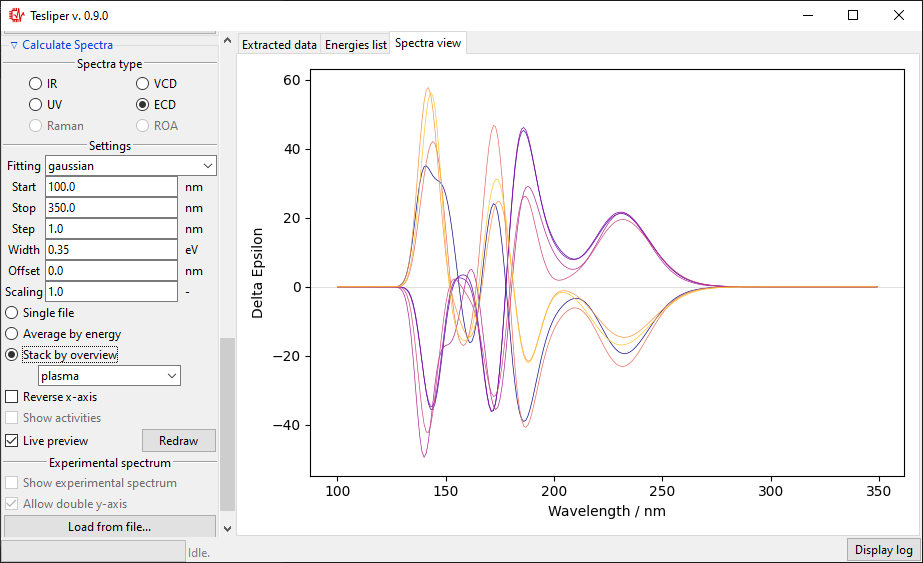
- "Spectra view" tab shows calculated spectra. It may be controlled using "Calculate spectra" section. After choosing a spectra genre to calculate you may control if it is simulated using lorentzian or gaussian fitting function, change peak width, spectra bounds, etc. You may view spectra for one conformer, all of them stacked, or averaged. You may also load an experimental spectrum (.txt or .csv format) for comparison.
Once done with extracting files and tweaking parameters, export selected data to desired format or save the session for later using buttons in "Session control" section.
A detailed tutorial with screenshots is available in the documentation: https://tesliper.readthedocs.io/en/stable/gui.html.
This project is licensed with BSD 2-Clause license. See LICENSE.txt file for details.
Contributions are welcome! tesliper is a growing project and definitely has room
for improvements.
Bug reports are of great value, if you encounter a problem please let me know by
submitting a new issue.
If you have a suggestion how tesliper can be improved, please let me know as well!
If you'd like to contribute to tesliper's codebase, that's even better! If there is a
specific bug that you know how to fix or a feature you'd like to develop, please let me
know via issues. To start coding, get your
working copy of tesliper's source code by cloning the repository and then setup your
environment by installing development dependencies (probably to a virtual environment):
python -m pip install .[dev]
Please remember to add/update relevant tests along with your code changes. Make sure the test suite passes by running
python -m pytest test
or, if you'd like to also generate the coverage report, you can invoke the pytest with the pytest-cov plugin (included in the "dev" installation), as shown below. The .coveragerc file includes plugin's configuration (i.e. to omit GUI code).
python -m pytest --cov=tesliper --cov-config=.coveragerc test
If you contribute code to tesliper, the test coverage badge will be updated by the CI
service after your changes are merged.
Although mypy is not incorporated in tesliper's
development (yet!), I believe that type hints greatly improve the experience of using
the package. Please, add them to the new code you submit. If you're willing to
supplement typing of the existing code, that would be very much welcome!
tesliper's codebase is formatted with black and
isort, please use these tools before submitting your
code contribution. To make it easier, you may use a pre-commit configuration available
in this repository. To include it in your workflow, simply run pre-commit install in
your copy's root directory. Note, that this configuration also sets up
flake8 linter.
To get your change introduced to the codebase, please make a Pull Request to the fixes
branch for quick bug fixes or to the dev branch for new features and bigger changes.
If at a loss, do not hesitate to reach to me directly! :)
Ideas for possible future improvements to the software are listed below. Based on the feedback from the Community, I will decide, which ones are desired and worth working on.
- command line interface
- support for Jaguar & other packages
- option for using
cclibfor parsing - spectra comparison feature
- velocity and length electronic spectra comparison
- parsing conformational search files
- GUI: manually choose spectra colour
- supplement logging
- auto finding optimal spectra range
- GUI: export spectra as image
- GUI: spectra colour by population
- GUI: drag&drop support
- GUI: display tooltips on hover
Many thanks to the scientists, who advised me on the domain-specific details and helped to test the software:
- Joanna Rode
0000-0003-0592-4053
- Magdalena Jawiczuk
0000-0003-2576-4042
- Marcin Górecki
0000-0001-7472-3875
as well as to people, who reviewed the project: @alejandrogallo and @arepstein.
I'm very happy to announce, that tesliper has been published in the Journal of Open
Source Software! If you find
tesliper useful in your research, please give it a credit by citing it:
- Więcław, M. M., (2022). tesliper: a theoretical spectroscopist's little helper. Journal of Open Source Software, 7(72), 4164, DOI: 10.21105/joss.04164
@article{tesliper2022,
doi = {10.21105/joss.04164},
url = {https://doi.org/10.21105/joss.04164},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {72},
pages = {4164},
author = {Michał M. Więcław},
title = {tesliper: a theoretical spectroscopist's little helper},
journal = {Journal of Open Source Software}
}



 0000-0003-0592-4053
0000-0003-0592-4053